Part 2: Hello Containers¶
In Part 1, you learned how to use the basic building blocks of Nextflow to assemble a simple pipeline capable of processing some text and parallelizing execution if there were multiple inputs.
However, you were limited to basic UNIX tools available in your environment. Real-world tasks often require various tools and packages not included by default. Typically, you'd need to install these tools, manage their dependencies, and resolve any conflicts.
That is all very tedious and annoying, so we're going to show you how to use containers to solve this problem much more conveniently.
Note
We'll be teaching this using the technology Docker, but Nextflow supports several other container technologies as well.
1. Use a container directly¶
A container is a lightweight, standalone, executable unit of software created from a container image that includes everything needed to run an application including code, system libraries and settings. To use a container you usually download or "pull" a container image from a container registry, and then run the container image to create a container instance.
1.1. Pull the container image¶
Let's pull a container image that contains the cowsay command so we can use it to display some text in a fun way.
1.2 Use the container to execute a single command¶
The docker run command is used to spin up a container instance from a container image and execute a command in it.
The --rm flag tells Docker to remove the container instance after the command has completed.
docker run --rm 'community.wave.seqera.io/library/pip_cowsay:131d6a1b707a8e65' cowsay -t "Hello World"
1.2. Spin up the container interactively¶
You can also run a container interactively, which will give you a shell prompt inside the container.
Notice that the prompt has changed to (base) root@b645838b3314:/tmp#, which indicates that you are now inside the container.
If we run:
(base) root@b645838b3314:/tmp# ls /
bin dev etc home lib media mnt opt proc root run sbin srv sys tmp usr var
You can see that the filesystem inside the container is different from the filesystem on your host system.
1.3. Run the command¶
Now that you are inside the container, you can run the cowsay command directly.
Tip
Us the '-c' flag to pick a different "cow" from this list:
beavis, cheese, cow, daemon, dragon, fox, ghostbusters, kitty, meow, miki, milk, octopus, pig, stegosaurus, stimpy, trex, turkey, turtle, tux
Output:
___________
| Hello World |
===========
\
\
\
.--.
|o_o |
|:_/ |
// \ \
(| | )
/'\_ _/`\
\___)=(___/
1.4. Exit the container¶
To exit the container, you can type exit at the prompt or use the Ctrl+D keyboard shortcut.
Your prompt should now be back to what it was before you started the container.
1.5. Mounting data into containers¶
When you run a container, it is isolated from the host system by default. This means that the container can't access any files on the host system unless you explicitly tell it to. One way to do this is to mount a volume from the host system into the container.
Prior to working on the next task, confirm that you are in the hello-nextflow directory. The last part of the path shown when you type pwd should be hello-nextflow.
Then run:
docker run --rm -it -v $(pwd)/containers/data:/data 'community.wave.seqera.io/library/pip_cowsay:131d6a1b707a8e65' /bin/bash
Let's explore the contents of the container.
Note that we need to navigate to the /data directory inside the container to see the contents of the data directory on the host system.
(base) root@08dd2d3efbd4:/tmp# ls
conda.yml environment.lock
(base) root@08dd2d3efbd4:/tmp# cd /data
(base) root@08dd2d3efbd4:/data# ls
greetings.csv pioneers.csv
1.6. Use the mounted data¶
Now that we have mounted the data directory into the container, we can use the cowsay command to display the contents of the greetings.csv file.
To do this we'll use the syntax -t "$(cat data/greetings.csv)" to output the contents of the file into the cowsay command.
Output:
__________________
| Hello,Bonjour,Holà |
==================
\
\
\
\
,.
(_|,.
,' /, )_______ _
__j o``-' `.'-)'
(") \'
`-j |
`-._( /
|_\ |--^. /
/_]'|_| /_)_/
/_]' /_]'
Now exit the container once again:
Takeaway¶
You know how to pull a container and run it interactively, make your data accessible to it, which lets you try commands without having to install any software on your system.
What's next?¶
Learn how to get a container image for any pip/conda-installable tool.
2. Use containers in Nextflow¶
Nextflow has built-in support for running processes inside containers to let you run tools you don't have installed in your compute environment. This means that you can use any container image you like to run your processes, and Nextflow will take care of pulling the image, mounting the data, and running the process inside it.
2.1. Add a container directive to your process¶
Edit the hello-containers.nf script to add a container directive to the cowsay process.
Before:
After:
process cowSay {
publishDir 'containers/results', mode: 'copy'
container 'community.wave.seqera.io/library/pip_cowsay:131d6a1b707a8e65'
2.2. Run Nextflow pipelines using containers¶
Run the script to see the container in action.
Note
The nextflow.config in our current working directory contains docker.enabled = true, which tells Nextflow to use Docker to run processes.
Without that configuration we would have to specify the -with-docker flag when running the script.
2.3. Check the results¶
You should see a new directory called containers/results that contains the output of the cowsay process.
_______
| Bonjour |
=======
\
\
^__^
(oo)\_______
(__)\ )\/\
||----w |
|| ||
2.4. Explore how Nextflow launched the containerized task¶
Let's take a look at the task directory for one of the cowsay tasks to see how Nextflow works with containers under the hood.
Check the output from your nextflow run command to find the task ID for the cowsay process.
Then check out the task directory for that task.
tree -a work/8c/738ac55b80e7b6170aa84a68412454
work/8c/738ac55b80e7b6170aa84a68412454
├── .command.begin
├── .command.err
├── .command.log
├── .command.out
├── .command.run
├── .command.sh
├── .exitcode
├── cowsay-output-Bonjour.txt
└── output-Bonjour.txt -> /workspace/gitpod/nf-training/hello-nextflow/work/0e/e96c123cb7ae9ff7b7bed1c5444009/output-Bonjour.txt
1 directory, 9 files
Open the .command.run file which holds all the busywork that Nextflow does under the hood.
Search for nxf_launch and you should see something like this:
nxf_launch() {
docker run -i --cpu-shares 1024 -e "NXF_TASK_WORKDIR" -v /workspace/gitpod/nf-training/hello-nextflow/work:/workspace/gitpod/nf-training/hello-nextflow/work -w "$NXF_TASK_WORKDIR" --name $NXF_BOXID community.wave.seqera.io/library/pip_cowsay:131d6a1b707a8e65 /bin/bash -ue /workspace/gitpod/nf-training/hello-nextflow/work/8c/738ac55b80e7b6170aa84a68412454/.command.sh
}
As you can see, Nextflow is using the docker run command to launch the task.
It also mounts the task's working directory into the container, sets the working directory inside the container to the task's working directory, and runs our templated bash script in the .command.sh file.
All the hard work we learned about in the previous sections is done for us by Nextflow!
Takeaway¶
You know how to use containers in Nextflow to run processes.
What's next?¶
You have everything you need to continue to the next chapter of this training series. Optionally, continue on to learn how to get container images for tools you want to use in your Nextflow pipelines.
3. Optional Topic: How to find or make container images¶
Some software developers provide container images for their software that are available on container registries like Docker Hub, but many do not. In this optional section, we'll show you to two ways to get a container image for tools you want to use in your Nextflow pipelines: using Seqera Containers and building the container image yourself.
You'll be getting/building a container image for the quote pip package, which will be used in the exercise at the end of this section.
3.1. Get a container image from Seqera Containers¶
Seqera Containers is a free service that builds container images for pip and conda (including bioconda) installable tools.
Navigate to Seqera Containers and search for the quote pip package.
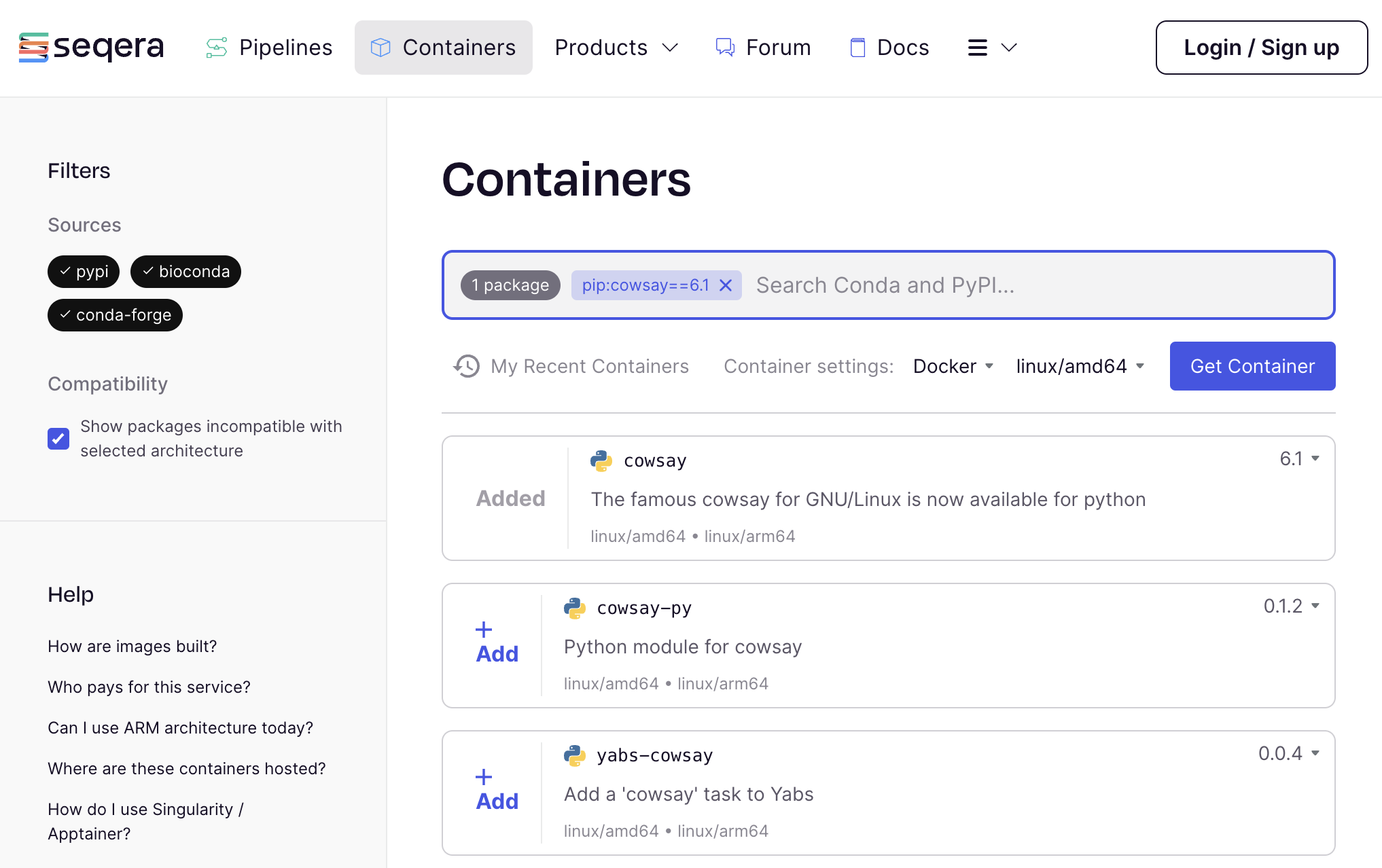
Click on "+Add" and then "Get Container" to request a container image for the quote pip package.
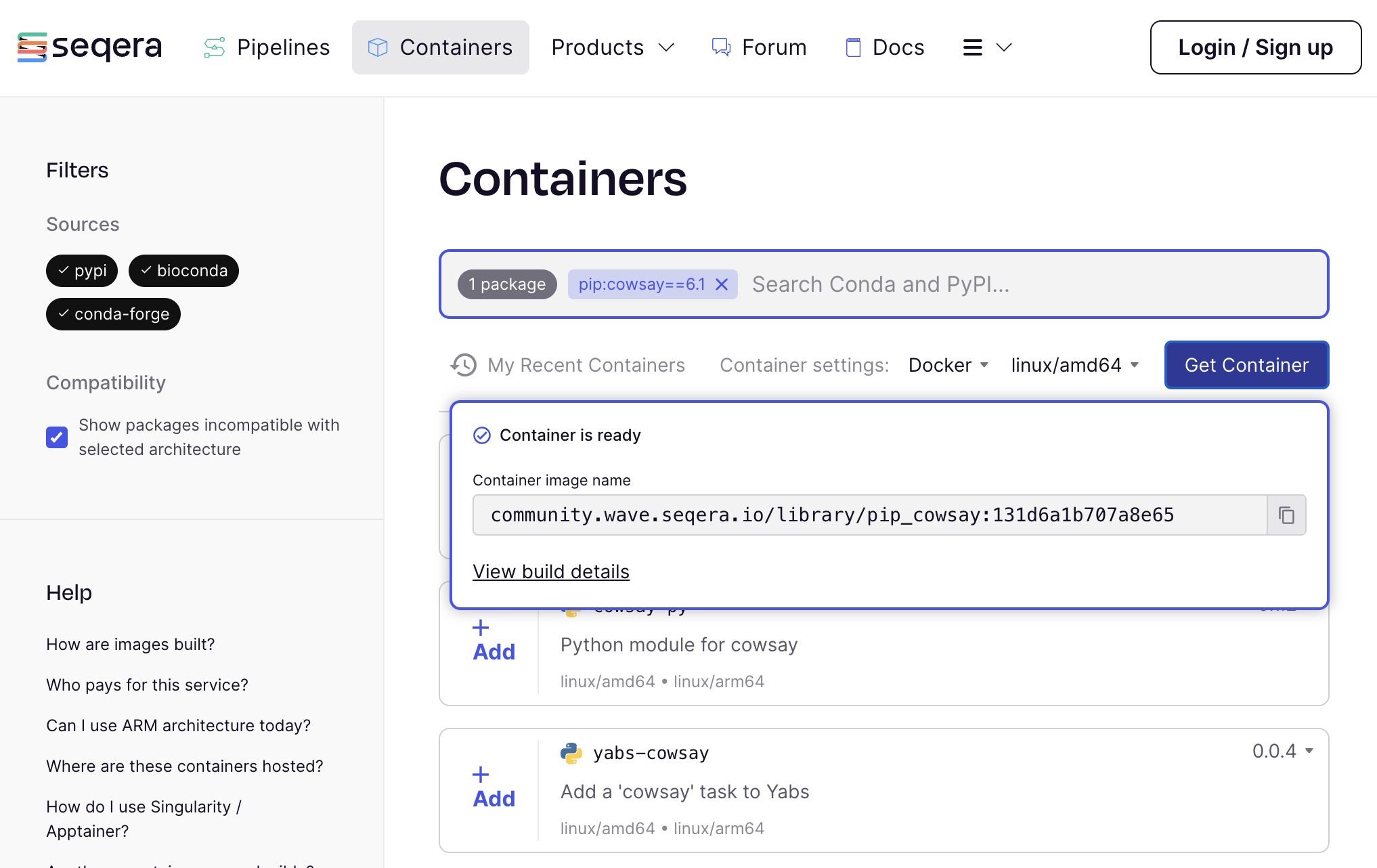
If this is the first time a community container has been built for this version of the package, it may take a few minutes to complete.
Click to copy the URI (e.g. community.wave.seqera.io/library/pip_quote:ae07804021465ee9) of the container image that was created for you.
You can now use the container image to run the quote command and get a random saying from Grace Hopper.
Output:
Humans are allergic to change. They love to say, 'We've always done it
this way.' I try to fight that. That's why I have a clock on my wall
that runs counter-clockwise.
3.2. Build the container image yourself¶
Let's use some build details from the Seqera Containers website to build the container image for the quote pip package ourselves.
Return to the Seqera Containers website and click on the "Build Details" button.
The first item we'll look at is the Dockerfile, a type of script file that contains all the commands needed to build the container image.
We've added some explanatory comments to the Dockerfile below to help you understand what each part does.
# Start from the micromamba base docker image
FROM mambaorg/micromamba:1.5.10-noble
# Copy the conda.yml file into the container
COPY --chown=$MAMBA_USER:$MAMBA_USER conda.yml /tmp/conda.yml
# Install various utilities for Nextflow to use and the packages in the conda.yml file
RUN micromamba install -y -n base -f /tmp/conda.yml \
&& micromamba install -y -n base conda-forge::procps-ng \
&& micromamba env export --name base --explicit > environment.lock \
&& echo ">> CONDA_LOCK_START" \
&& cat environment.lock \
&& echo "<< CONDA_LOCK_END" \
&& micromamba clean -a -y
# Run the container as the root user
USER root
# Set the PATH environment variable to include the micromamba installation directory
ENV PATH="$MAMBA_ROOT_PREFIX/bin:$PATH"
The second item we'll look at is the conda.yml file, which contains the list of packages that need to be installed in the container image.
Copy the contents of these files into the stubs located in the containers/build directory, then run the following command to build the container image yourself.
Note
We use the -t quote:latest flag to tag the container image with the name quote and the tag latest.
We will be able to use this tag to refer to the container image when running it on this system.
After it has finished building, you can run the container image you just built.
Takeaway¶
You've learned two different ways to get a container image for a tool you want to use in your Nextflow pipelines: using Seqera Containers and building the container image yourself.
What's next?¶
You have everything you need to continue to the next chapter of this training series.
You can also continue on with an optional exercise to fetch quotes on computer/biology pioneers using the quote container and output them using the cowsay container.
4. Bonus Exercise: Make the cow quote famous scientists¶
This section contains some stretch exercises, to practice what you've learned so far. Doing these exercises is not required to understand later parts of the training, but provide a fun way to reinforce your learnings by figuring out how to make the cow quote famous scientists.
_________________________________________________
/ \
| Humans are allergic to change. They love to |
| say, 'We've always done it this way.' I try to fi |
| ght that. That's why I have a clock on my wall th |
| at runs counter-clockwise. |
| -Grace Hopper |
\ /
=================================================
\
\
^__^
(oo)\_______
(__)\ )\/\
||----w |
|| ||
4.1. Modify the hello-containers.nf script to use a getQuote process¶
We have a list of computer and biology pioneers in the containers/data/pioneers.csv file.
At a high level, to complete this exercise you will need to:
- Modify the default
params.input_fileto point to thepioneers.csvfile. - Create a
getQuoteprocess that uses thequotecontainer to fetch a quote for each input. - Connect the output of the
getQuoteprocess to thecowsayprocess to display the quote.
For the quote container image, you can either use the one you built yourself in the previous stretch exercise or use the one you got from Seqera Containers .
Hint
A good choice for the script block of your getQuote process might be:
You can find a solution to this exercise in containers/solutions/hello-containers-4.1.nf.
4.2. Modify your Nextflow pipeline to allow it to execute in quote and sayHello modes.¶
Add some branching logic using to your pipeline to allow it to accept inputs intended for both quote and sayHello.
Here's an example of how to use an if statement in a Nextflow workflow:
Hint
You can use new_ch = processName.out to assign a name to the output channel of a process.
You can find a solution to this exercise in containers/solutions/hello-containers-4.2.nf.
Takeaway¶
You know how to use containers in Nextflow to run processes, and how to build some branching logic into your pipelines!
What's next?¶
Celebrate, take a stretch break and drink some water!
When you are ready, move on to Part 3 of this training series to learn how to apply what you've learned so far to a more realistic data analysis use case.