Part 2: Hello GATK¶
The GATK (Genome Analysis Toolkit) is a widely used software package developed by the Broad Institute to analyze high-throughput sequencing data. We're going to use GATK and a related tool, Samtools, in a very basic pipeline that identifies genomic variants through a method called variant calling.
Note
Don't worry if you're not familiar with GATK or genomics in general. We'll summarize the necessary concepts as we go, and the workflow implementation principles we demonstrate here apply broadly to any command line tool that takes in some input files and produce some output files.
A full variant calling pipeline typically involves a lot of steps. For simplicity, we are only going to look at the core variant calling steps.
Method overview¶
- Generate an index file for each BAM input file using Samtools
- Run the GATK HaplotypeCaller on each BAM input file to generate per-sample variant calls in GVCF (Genomic Variant Call Format)
Dataset¶
- A reference genome consisting of the human chromosome 20 (from hg19/b37) and its accessory files (index and sequence dictionary). The reference files are compressed to keep the Gitpod size small so we'll have to decompress them in order to use them.
- Three whole genome sequencing samples corresponding to a family trio (mother, father and son), which have been subset to a small portion on chromosome 20 to keep the file sizes small. The sequencing data is in BAM (Binary Alignment Map) format, i.e. genome sequencing reads that have already been mapped to the reference genome.
- A list of genomic intervals, i.e. coordinates on the genome where our samples have data suitable for calling variants.
0. Warmup: Run Samtools and GATK directly¶
Just like in the Hello World example, we want to try out the commands manually before we attempt to wrap them in a workflow. The difference here is that we're going to use Docker containers to obtain and run the tools.
0.1. Index a BAM input file with Samtools¶
0.1.1. Pull the samtools container¶
0.1.2. Spin up the container interactively¶
0.1.3. Run the indexing command¶
0.1.4. Check that the BAM index has been produced¶
This should show:
Where reads_mother.bam.bai has been created as an index to reads_mother.bam.
0.1.5. Exit the container¶
0.2. Call variants with GATK HaplotypeCaller¶
0.2.1. Decompress the reference genome files¶
0.2.2. Pull the GATK container¶
0.2.3. Spin up the container interactively¶
0.2.4. Run the variant calling command¶
gatk HaplotypeCaller \
-R /data/ref/ref.fasta \
-I /data/bam/reads_mother.bam \
-O reads_mother.g.vcf \
-L /data/intervals.list \
-ERC GVCF
0.2.5. Check the contents of the output file¶
1. Write a single-stage workflow that runs Samtools index on a BAM file¶
1.1. Define the indexing process¶
/*
* Generate BAM index file
*/
process SAMTOOLS_INDEX {
container 'quay.io/biocontainers/samtools:1.19.2--h50ea8bc_1'
input:
path input_bam
output:
path "${input_bam}.bai"
"""
samtools index '$input_bam'
"""
}
1.2. Add parameter declarations up top¶
/*
* Pipeline parameters
*/
// Execution environment setup
params.baseDir = "/workspace/gitpod/hello-nextflow"
$baseDir = params.baseDir
// Primary input
params.reads_bam = "${baseDir}/data/bam/reads_mother.bam"
1.3. Add workflow block to run SAMTOOLS_INDEX¶
workflow {
// Create input channel (single file via CLI parameter)
reads_ch = Channel.from(params.reads_bam)
// Create index file for input BAM file
SAMTOOLS_INDEX(reads_ch)
}
1.4. Run it to verify you can run the indexing step¶
Should produce something like:
N E X T F L O W ~ version 23.10.1
Launching `hello-gatk.nf` [compassionate_cray] DSL2 - revision: 9b97744397
executor > local (1)
[bf/072bd7] process > SAMTOOLS_INDEX (1) [100%] 1 of 1 ✔
Takeaway¶
You know how to wrap a real bioinformatics tool in a single-step Nextflow workflow.
What's next?¶
Add a second step that consumes the output of the first.
2. Add a second step that runs GATK HaplotypeCaller on the indexed BAM file¶
2.1. Define the variant calling process¶
/*
* Call variants with GATK HapolotypeCaller in GVCF mode
*/
process GATK_HAPLOTYPECALLER {
container "broadinstitute/gatk:4.5.0.0"
input:
path input_bam
path input_bam_index
path ref_fasta
path ref_index
path ref_dict
path interval_list
output:
path "${input_bam}.g.vcf"
path "${input_bam}.g.vcf.idx"
"""
gatk HaplotypeCaller \
-R ${ref_fasta} \
-I ${input_bam} \
-O ${input_bam}.g.vcf \
-L ${interval_list} \
-ERC GVCF
"""
}
2.2. Add accessory inputs up top¶
// Accessory files
params.genome_reference = "${baseDir}/data/ref/ref.fasta"
params.genome_reference_index = "${baseDir}/data/ref/ref.fasta.fai"
params.genome_reference_dict = "${baseDir}/data/ref/ref.dict"
params.calling_intervals = "${baseDir}/data/intervals.list"
2.3. Add a call to the workflow block to run GATK_HAPLOTYPECALLER¶
// Call variants from the indexed BAM file
GATK_HAPLOTYPECALLER(
reads_ch,
SAMTOOLS_INDEX.out,
params.genome_reference,
params.genome_reference_index,
params.genome_reference_dict,
params.calling_intervals
)
2.4. Run the workflow to verify that the variant calling step works¶
Now we see the two processes being run:
N E X T F L O W ~ version 23.10.1
Launching `hello-gatk.nf` [lethal_keller] DSL2 - revision: 30a64b9325
executor > local (2)
[97/0f85bf] process > SAMTOOLS_INDEX (1) [100%] 1 of 1 ✔
[2d/43c247] process > GATK_HAPLOTYPECALLER (1) [100%] 1 of 1 ✔
If you check the work directory, you'll find the output file reads_mother.bam.g.vcf. Because this is a small test file, you can click on it to open it and view the contents, which consist of 92 lines of header metadata followed by a list of genomic variant calls, one per line.
Note
A GVCF is a special kind of VCF that contains non-variant records as well as variant calls. The first actual variant call in this file occurs at line 325:
Takeaway¶
You know how to make a very basic two-step variant calling workflow.
What's next?¶
Make the workflow handle multiple samples in bulk.
3. Adapt the workflow to run on a batch of samples¶
3.1. Turn the input param declaration into a list of the three samples¶
// Primary input
params.reads_bam = [
"${baseDir}/data/bam/reads_mother.bam",
"${baseDir}/data/bam/reads_father.bam",
"${baseDir}/data/bam/reads_son.bam"
]
3.2. Run the workflow to verify that it runs on all three samples¶
Uh-oh! It fails with an error like this:
A USER ERROR has occurred: Traversal by intervals was requested but some input files are not indexed.
Please index all input files:
samtools index reads_son.bam
This is because the file paths are different for the BAM files and their index files, so GATK does not recognize that they go together. This can be addressed by passing in the index files explicitly, but there's a plot twist: the script as written so far is not safe for running on multiple samples, because the order of outputs is not guaranteed. Even if we solved the indexing problem, we would end up with race condition issues. So we need to make sure the BAM files and their index files travel together through the channels.
3.3. Change the output of the SAMTOOLS_INDEX process into a tuple that keeps the input file and its index together¶
Before:
After:
3.4. Change the input to the GATK_HAPLOTYPECALLER process to be a tuple¶
Before:
After:
3.5. Update the call to GATK_HAPLOTYPECALLER in the workflow block¶
Before:
After:
3.6. Run the workflow to verify it works correctly on all three samples now¶
This time everything should run correctly:
N E X T F L O W ~ version 23.10.1
Launching `hello-gatk.nf` [adoring_hopper] DSL2 - revision: 8cad21ea51
[e0/bbd6ef] Submitted process > SAMTOOLS_INDEX (3)
[71/d26b2c] Submitted process > SAMTOOLS_INDEX (2)
[e6/6cad6d] Submitted process > SAMTOOLS_INDEX (1)
[26/73dac1] Submitted process > GATK_HAPLOTYPECALLER (1)
[23/12ed10] Submitted process > GATK_HAPLOTYPECALLER (2)
[be/c4a067] Submitted process > GATK_HAPLOTYPECALLER (3)
Takeaway¶
You know how to make a variant calling workflow run on multiple samples (independently).
What's next?¶
Make it easier to handle samples in bulk.
4. Make it nicer to run on arbitrary samples by using a list of files as input¶
4.1. Create a text file listing the input paths¶
/workspace/gitpod/hello-nextflow/data/bam/reads_mother.bam
/workspace/gitpod/hello-nextflow/data/bam/reads_father.bam
/workspace/gitpod/hello-nextflow/data/bam/reads_son.bam
4.2. Update the parameter default¶
Before:
// Primary input
params.reads_bam = [
"${baseDir}/data/bam/reads_mother.bam",
"${baseDir}/data/bam/reads_father.bam",
"${baseDir}/data/bam/reads_son.bam"
]
After:
// Primary input (list of input files, one per line)
params.reads_bam = "${baseDir}/data/sample_bams.txt"
4.3. Update the channel factory to read lines from a file¶
Before:
After:
// Create input channel from list of input files in plain text
reads_ch = Channel.fromPath(params.reads_bam).splitText()
4.4. Run the workflow to verify that it works correctly¶
This should produce essentially the same result as before:
N E X T F L O W ~ version 23.10.1
Launching `hello-gatk.nf` [kickass_faggin] DSL2 - revision: dcfa9f34e3
[ff/0c08e6] Submitted process > SAMTOOLS_INDEX (2)
[75/bcae76] Submitted process > SAMTOOLS_INDEX (1)
[df/75d25a] Submitted process > SAMTOOLS_INDEX (3)
[00/295d75] Submitted process > GATK_HAPLOTYPECALLER (1)
[06/89c1d1] Submitted process > GATK_HAPLOTYPECALLER (2)
[58/866482] Submitted process > GATK_HAPLOTYPECALLER (3)
Takeaway¶
You know how to make a variant calling workflow handle a list of input samples.
What's next?¶
Turn the list of input files into a samplesheet by including some metadata.
5. Upgrade to using a (primitive) samplesheet¶
This is a very common pattern in Nextflow pipelines.
5.1. Add a header line and the sample IDs to a copy of the sample list, in CSV format¶
ID,reads_bam
NA12878,/workspace/gitpod/hello-nextflow/data/bam/reads_mother.bam
NA12877,/workspace/gitpod/hello-nextflow/data/bam/reads_father.bam
NA12882,/workspace/gitpod/hello-nextflow/data/bam/reads_son.bam
5.2. Update the parameter default¶
Before:
// Primary input (list of input files, one sample per line)
params.reads_bam = "${baseDir}/data/bam/sample_bams.txt"
After:
// Primary input (samplesheet in CSV format with ID and file path, one sample per line)
params.reads_bam = "${baseDir}/data/samplesheet.csv"
5.3. Update the channel factory to parse a CSV file¶
Before:
// Create input channel from list of input files in plain text
reads_ch = Channel.fromPath(params.reads_bam).splitText()
After:
// Create input channel from samplesheet in CSV format
reads_ch = Channel.fromPath(params.reads_bam)
.splitCsv(header: true)
.map{row -> [row.id, file(row.reads_bam)]}
5.4. Add the sample ID to the SAMTOOLS_INDEX input definition¶
Before:
After:
5.5. Run the workflow to verify that it works¶
If everything is wired up correctly, it should produce essentially the same result.
N E X T F L O W ~ version 23.10.1
Launching `hello-gatk.nf` [extravagant_panini] DSL2 - revision: 56accbf948
[19/00f4a5] Submitted process > SAMTOOLS_INDEX (3)
[4d/532d60] Submitted process > SAMTOOLS_INDEX (1)
[08/5628d6] Submitted process > SAMTOOLS_INDEX (2)
[18/21a0ae] Submitted process > GATK_HAPLOTYPECALLER (1)
[f0/4e8155] Submitted process > GATK_HAPLOTYPECALLER (2)
[d5/73e1c4] Submitted process > GATK_HAPLOTYPECALLER (3)
Takeaway¶
You know how to make a variant calling workflow handle a basic samplesheet.
What's next?¶
Add a joint genotyping step that combines the data from all the samples.
6. Stretch goal: Add joint genotyping step¶
To complicate matters a little, the GATK variant calling method calls for a consolidation step where we combine and re-analyze the variant calls obtained per sample in order to obtain definitive 'joint' variant calls for a group or cohort of samples (in this case, the family trio).
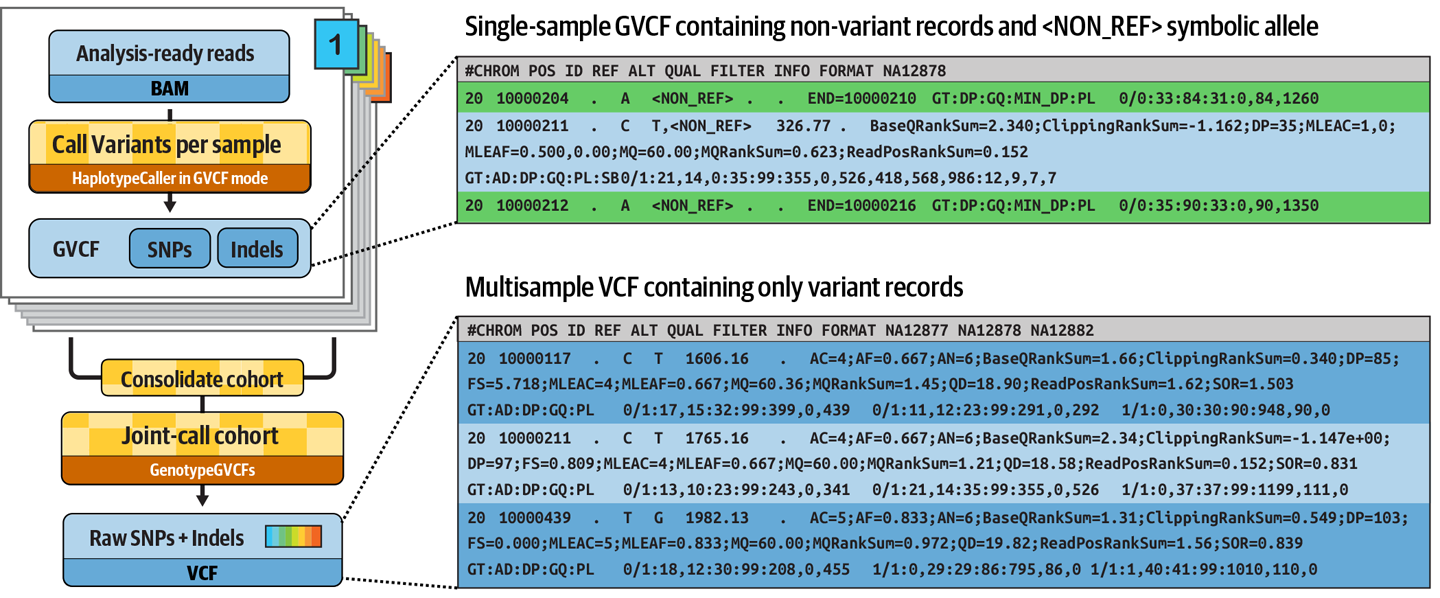
This involves using a GATK tool called GenomicsDBImport that combines the per-sample calls into a sort of mini-database, followed by another GATK tool, GenotypeGVCFs, which performs the actual 'joint genotyping' analysis. These two tools can be run in series within the same process.
One slight complication is that these tools require the use of a sample map that lists per-sample GVCF files, which is different enough from a samplesheet that we need to generate it separately. And for that, we need to pass the sample ID between processes.
Tip
For a more sophisticated and efficient method of metadata propagation, see the topic of meta maps.
6.2. Add the sample ID to the tuple emitted by SAMTOOLS_INDEX¶
Before:
After:
6.3. Add the sample ID to the GATK_HAPLOTYPECALLER process input and output definitions¶
Before:
input:
tuple path(input_bam), path(input_bam_index)
...
output:
path "${input_bam}.g.vcf"
path "${input_bam}.g.vcf.idx"
After:
input:
tuple val(id), path(input_bam), path(input_bam_index)
...
output:
tuple val(id), path("${input_bam}.g.vcf"), path("${input_bam}.g.vcf.idx")
6.4. Generate a sample map based on the output of GATK_HAPLOTYPECALLER¶
// Create a sample map of the output GVCFs
sample_map = GATK_HAPLOTYPECALLER.out.collectFile(){ id, gvcf, idx ->
["${params.cohort_name}_map.tsv", "${id}\t${gvcf}\t${idx}\n"]
}
6.5. Write a process that wraps GenomicsDBImport and GenotypeGVCFs called GATK_JOINTGENOTYPING¶
/*
* Consolidate GVCFs and apply joint genotyping analysis
*/
process GATK_JOINTGENOTYPING {
container "broadinstitute/gatk:4.5.0.0"
input:
path(sample_map)
val(cohort_name)
path ref_fasta
path ref_index
path ref_dict
path interval_list
output:
path "${cohort_name}.joint.vcf"
path "${cohort_name}.joint.vcf.idx"
"""
gatk GenomicsDBImport \
--sample-name-map ${sample_map} \
--genomicsdb-workspace-path ${cohort_name}_gdb \
-L ${interval_list}
gatk GenotypeGVCFs \
-R ${ref_fasta} \
-V gendb://${cohort_name}_gdb \
-O ${cohort_name}.joint.vcf \
-L ${interval_list}
"""
}
6.6. Add call to workflow block to run GATK_JOINTGENOTYPING¶
// Consolidate GVCFs and apply joint genotyping analysis
GATK_JOINTGENOTYPING(
sample_map,
params.cohort_name,
params.genome_reference,
params.genome_reference_index,
params.genome_reference_dict,
params.calling_intervals
)
6.7. Add default value for the cohort name parameter up top¶
6.8. Run the workflow to verify that it generates the final VCF output as expected¶
Now we see the additional process show up in the log output (showing the compact view):
N E X T F L O W ~ version 23.10.1
Launching `hello-gatk.nf` [nauseous_thompson] DSL2 - revision: b346a53aae
executor > local (7)
[d1/43979a] process > SAMTOOLS_INDEX (2) [100%] 3 of 3 ✔
[20/247592] process > GATK_HAPLOTYPECALLER (3) [100%] 3 of 3 ✔
[14/7145b6] process > GATK_JOINTGENOTYPING (1) [100%] 1 of 1 ✔
You can find the final output file, family_trio.joint.vcf, in the work directory for the last process. Click on it to open it and you'll see 40 lines of metadata header followed by just under 30 jointly genotyped variant records (meaning at least one of the family members has a variant genotype at each genomic position listed).
Tip
Keep in mind the data files covered only a tiny portion of chromosome 20; the real size of a variant callset would be counted in millions of variants. That's why we use only tiny subsets of data for training purposes!
Takeaway¶
You know how to make a joint variant calling workflow that outputs a cohort VCF.
What's next?¶
Celebrate your success and take an extra long break! This was tough and you deserve it.
In future trainings, you'll learn more sophisticated methods for managing inputs and outputs (including using the publishDir directive to save the outputs you care about to a storage directory).
Good luck!